A number of methods to generate additional data for model training are available in the Generate Data tool. It is available via the main tool menu in the View tool:
The most common methods are available on the main Data Generation tab:
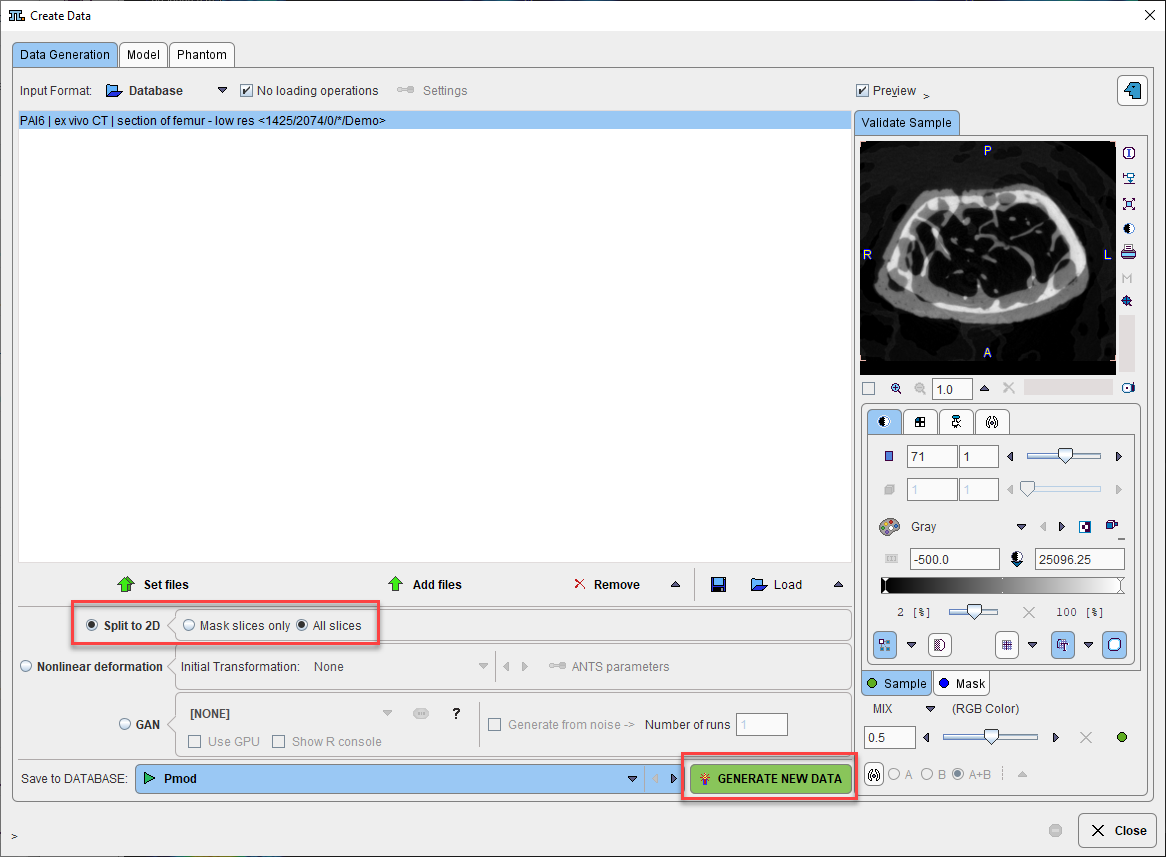
Input files can be specified using the typical Set Files interface.
Split to 2D is useful to prepare individual 2D slices from a 3D dataset for training of a model that should work slicewise. This method can be tested and understood using the PAI6 mouse microCT section of femur example in our Demo database. The 142 slice low-resolution series can be taken from the Demo database and split into 142 individual 2D series into the Pmod database, ready to start a Learning Set. If a reference segmentation result (SEGMENT mask) is available and Associated with the input series, pairs of Associated image/mask will be generated.
Nonlinear deformation takes advantage of the iterative ANTS elastic matching method to save all iterations between two input images.
GAN uses Generative Adversarial Networks to generate entirely novel image data. For example, this may be an image representing typical cardiac MR function data, starting from automatically generated noise input images.
We strongly recommend the use of our Database functionality for input/output to take advantage of the Association functionality and create data that can be rapidly incorporated into Learning Sets.
4D data can also be generated using a geometric model and simulated data through the Model and Phantom tabs. Please see the documentation for our Geometric Models tool (PGEM), section Generation of Dynamic PET Phantom Images, for more information and contact us for further assistance.